OpenLineage
The OpenLineage integration consumes OpenLineage metadata from various systems.
Grai offers detailed instructions for a number of common OpenLineage producers including Airflow.
Web App
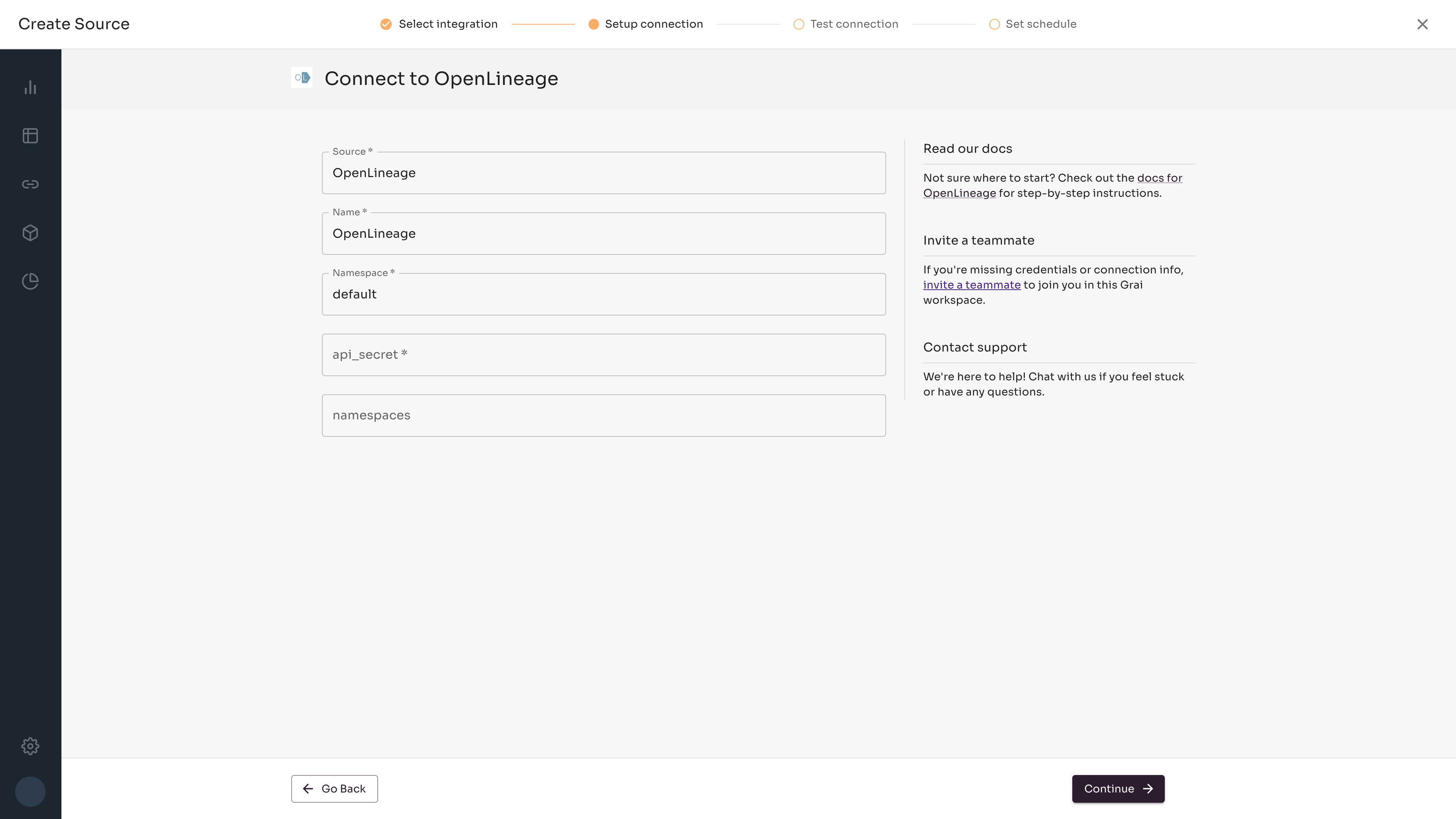
Fields
| Field | Value | Example |
|---|---|---|
| source | The name of the source, see sources | my-source |
| Name | Name for connection | OpenLineage |
| Namespace | Namespace for the connection, see namespaces | default |
| namespaces | Optional |
Webhook
In order to authenticate your webhook, you will need to create an API key in the Grai web app.
Grai will open an endpoint at /api/v1/openlineage/<connection_id>/ which can be used to receive OpenLineage events. You should point your OpenLineage clients to this endpoint.
See the OpenLineage docs (opens in a new tab) for how to configure your client to send events to a webhook.
An example config:
transport:
type: "http"
url: "https://api.grai.io"
endpoint: "api/v1/openlineage/<connection_id>/"
auth:
type: "api_key"
api_key: "<your_grai_api_key>"Python Library
The OpenLineage integration can be run as a standalone python library to convert OpenLineage events into Grai objects.
The library is available via pip
pip install grai_source_openlineageMore information about the API is available here.
Example
The library is split into a few distinct functions but if you only wish to extract nodes/edges you can do so as follows:
from grai_source_openlineage import OpenLineageIntegration
from grai_schemas.v1.source import SourceV1
source = SourceV1(name="my-source", type="my-type")
openlineage_params = {
"namespaces": {}
}
integration = OpenLineageIntegration(source=source, namespace="openlineage", **openlineage_params)
nodes, edges = integration.get_nodes_and_edges()In this case, we are putting all nodes and edges produced by OpenLineage in a single namespace.
In practice you usually don't want to do this because it will result in overlapping id's.
For example, an OpenLineage connection copying data from a source table my_table to a destination table my_table will
result in two nodes with the same id.
To avoid this, you can pass a namespaces parameter to the OpenLineageIntegration constructor which will map OpenLineage
namespaces to source and destination Grai namespaces.
namespaces = {<openlineage_namespace>: <grai_namepsace>}
integration = OpenLineageIntegration(source=source, namespaces=namespaces)